|
 |
 |

|

|
|
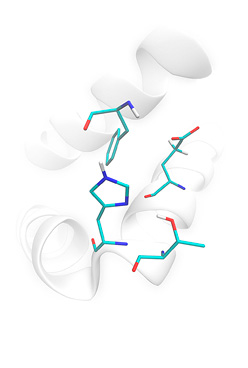
Preparation of cytochrome CYP450 3A4 from crude pdb file (pdb id 1w0f): missing (not resolved) residues were automatically build; orientation of Asn, Gln His side chains was optimized; ionization states of residues were fitted for given pH(7.0); positions of flexible hydrogen atoms were energy — optimized. You can download examples of protein structure preparation using Lead-Finder. Each example (archive file) contains description of the task and necessary structure files.
1pro: HIV-protease structure model
|
Protein structure preparationPreparing high quality model of a protein structure for computational chemistry studies starts with addition of hydrogen atoms to PDB coordinates of heavy atoms implying correct assignment of ionization states of protein residues and optimization of proton positions, which is a challenging scientific task. Lead-Finder saves user’s time and reduces expenses by including protein structure preparation tools that allow the:
Lead-Finder uses quite sophisticated theoretical approaches to assign optimal ionization states of protein residues at arbitrary pH, which is based on the recently introduced screened coulomb potential (SCP) model. SCP theory accounts for the dependence of electrostatic interactions between charged particles on such physicochemical properties of their microenvironment as hydrophilicity and degree of solvent exposure. Description of the SCP model can be found in original publications 1,2 , and details of its implementation in Lead-Finder program are given in the Technology section The quality of Lead-Finder predictions of ionization properties of proteins was validated by calculating pKa values of 100 residues from 15 proteins, for which robust experimental data were available. Lead-Finder demonstrated impressive results, yielding RMSD of predicted vs experimental pKa values of 0.7 for Asp and Glu residues, 0.8 for His residues, 0.85 for Lys residues, 1.25 for Tyr residues. Details of benchmarking experiments can be found in section Accuracy of pKa predictions.  |
 |
 |