|
 |
 |

|

|
|
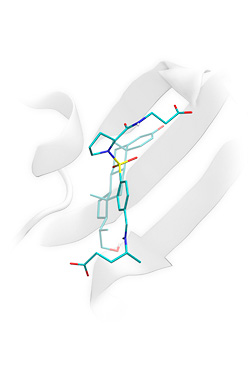
True active ligand of thymidine kinase correctly docked to the protein binding site and highly scored by Lead-Finder (opaque licorice) and low-scored inactive compound (semi-transparent licorice). You can download example of a virtual ligand screening study using Lead-Finder. Example (archive file) contains description of the task and necessary files (structure of the protein target, active and decoy ligands).
Virtual screening study of Peroxisome
|
Virtual screeningLead-Finder can perform virtual screening of libraries of chemical compounds to find a potent binder for a given target protein. During virtual screening each ligand from a library is docked to a target protein, and ligands are rank-ordered according to their binding potencies. Special type of scoring function (called VS-score) was implemented in Lead-Finder to rank ligands according to their activity. This scoring function combines the same energy contributions as other scoring functions implemented in Lead-Finder (used for pose ranking during the docking run and binding energy estimation for the docked ligand poses; see Technology section for details), however energy scaling coefficients were specially adjusted to attain better enrichment indicators during virtual screening experiments. Efficiency of Lead-Finder in virtual screening studies was extensively benchmarked on the set of 34 therapeutically relevant protein targets. As can be seen from corresponding benchmarking section Lead-Finder was able to achieve impressive enrichments for almost all protein targets. To make virtual screening of massive libraries feasible, special settings of the docking algorithm were designed to comprise the so-called screening regime, which is 2-4 times faster than the default docking regime. The fast screening mode retains almost the same accuracy of free energy of binding prediction and docking success rate as the default docking regime (see Speed of calculations section).  |
 |
 |